Introduction
BioBundle was a medium-difficulty reverse engineering CTF challenge from HackTheBox University CTF 2023: Brains & Bytes.
Tools
| Tool | Description |
|---|---|
| Ghidra | Ghidra is a software reverse engineering framework |
| Strace | strace is a diagnostic, debugging and instructional userspace utility for Linux. |
| Cyberchef | The Cyber Swiss Army Knife |
BioBundle
This challenge provided us with a binary that we could run and send some input.
┌──(kali㉿kali)-[~/Desktop/HTB-University/rev_biobundle]
└─$ file biobundle
biobundle: ELF 64-bit LSB pie executable, x86-64, version 1 (SYSV), dynamically linked, interpreter /lib64/ld-linux-x86-64.so.2, BuildID[sha1]=0ff6e374d4953a88af400e872f4df8d48fa5472e, for GNU/Linux 3.2.0, not stripped
┌──(kali㉿kali)-[~/Desktop/HTB-University/rev_biobundle]
└─$ ./biobundle
testinput
[x] Critical Failure
Opening it up in Ghidra, we can see two functions, main() and get_handle().
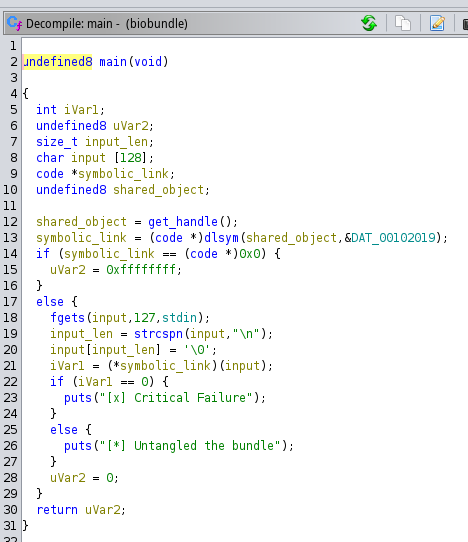
The main() function is pretty straightforward it has a symbolic link created with the value returned from the get_handle() function. If it can’t create the symbolic link, the function returns and finishes, but if it can, it will ask the user for input, which will be compared with the value from the symbolic link. If they are different, the program will throw the message “Critical Failure” but if it matches, the message “Untangled the bundle” will appear.
The main() function can be seen in the following image.
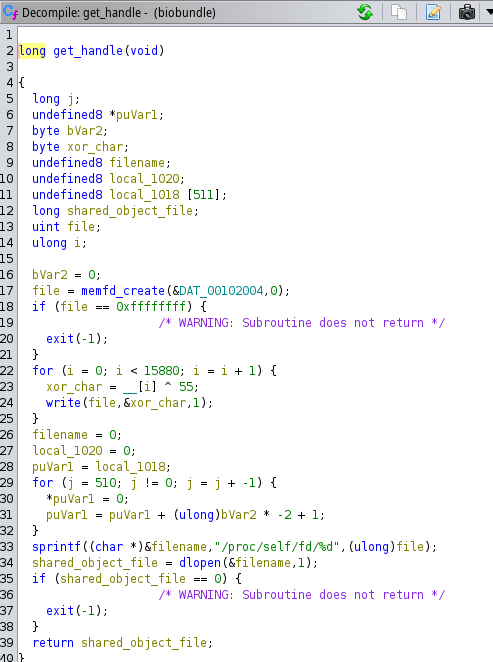
Moving on with the get_handle() function, here it starts by utilizing the memfd_create() function to create a file in memory. Then it will write the file contents char by char, and open the file has a shared object with the dlopen() at the location "/proc/self/fd/<filename>" finally return the handle of this object to the main function.
The get_handle() function can be seen in the following image.
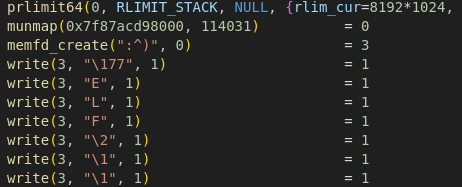
Now that we know what the program does, we can start dynamically analyzing it using strace and leaving the application running without inserting any input.
┌──(kali㉿kali)-[~/Desktop/HTB-University/rev_biobundle]
└─$ strace --output result ./biobundle
Opening the result file and searching for the memfd_create() call, it’s possible to see that there is an ELF file created in memory with the name ":^)".
We can now search for the process PID with the ps aux command.
┌──(kali㉿kali)-[~/Desktop/HTB-University/rev_biobundle]
└─$ ps aux
kali 125331 3.8 0.0 13780 7432 pts/0 Ss 12:11 0:00 /usr/bin/zsh
kali 125548 0.0 0.0 2516 1280 pts/0 S+ 12:12 0:00 ./biobundle
kali 125573 0.0 0.0 10976 4352 pts/10 R+ 12:12 0:00 ps aux
With the PID obtained, we can search for the file.
┌──(kali㉿kali)-[~/Desktop/HTB-University/rev_biobundle]
└─$ ls -lha /proc/125548/fd
total 0
dr-x------ 2 kali kali 4 Dec 8 12:12 .
dr-xr-xr-x 9 kali kali 0 Dec 8 12:12 ..
lrwx------ 1 kali kali 64 Dec 8 12:13 0 -> /dev/pts/0
lrwx------ 1 kali kali 64 Dec 8 12:13 1 -> /dev/pts/0
lrwx------ 1 kali kali 64 Dec 8 12:13 2 -> /dev/pts/0
lrwx------ 1 kali kali 64 Dec 8 12:12 3 -> '/memfd::^) (deleted)'
And there it is. Now, we can extract its contents to a new file. And open it in ghidra.
┌──(kali㉿kali)-[~/Desktop/HTB-University/rev_biobundle]
└─$ cat /proc/125548/fd/3 > extracted.elf
┌──(kali㉿kali)-[~/Desktop/HTB-University/rev_biobundle]
└─$ file extracted.elf
extracted.elf: ELF 64-bit LSB shared object, x86-64, version 1 (SYSV), dynamically linked, BuildID[sha1]=be4e99d76bd029ec4daa2deb1edd76a012797711, not stripped
This program will take an input and compare it with the stored value, as seen in the following image.
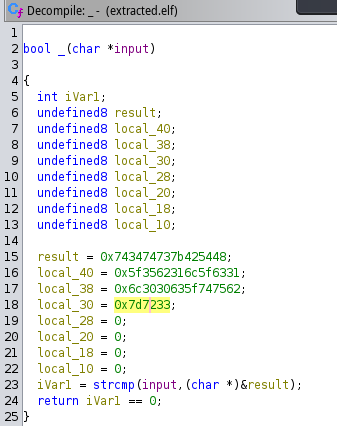
Now grabbing all hexadecimal values we can put this into cyberchef and decode it.
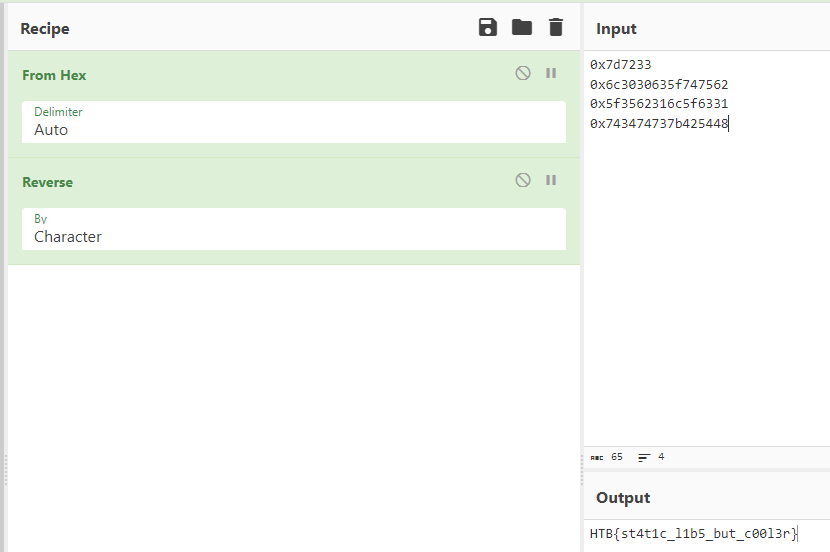
Giving us the flag HTB{st4t1c_l1b5_but_c00l3r}
Conclusion
To wrap things up this challenge provided an enjoyable opportunity to revisit some reverse engineering concepts.
Hope you liked this post.
Best Regards, Diogo.